Olfactory receptor neuron
| Olfactory receptor neuron | |
|---|---|
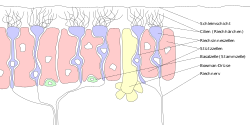 Labels in German. "Zellen" = "cell", "riech" = "smell", "Riechnerv" = olfactory nerve, "cillien" = cilia. | |
| Details | |
| System | Smell |
| Location | Olfactory epithelium in the nose |
| Shape | Bipolar sensory receptor |
| Function | Detect traces of chemicals in inhaled air (sense of smell) |
| Neurotransmitter | Glutamate[1] |
| Presynaptic connections | None |
| Postsynaptic connections | Olfactory bulb |
| Identifiers | |
| MeSH | D018034 |
| NeuroLex ID | nifext_116 |
| TH | H3.11.07.0.01003 |
| FMA | 67860 |
| Anatomical terms of neuroanatomy | |
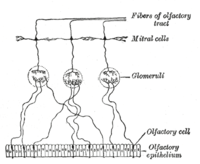

An olfactory receptor neuron (ORN), also called an olfactory sensory neuron (OSN), is a sensory neuron within the olfactory system.[2]
Structure
[edit]Humans have between 10 and 20 million olfactory receptor neurons (ORNs).[3] In vertebrates, ORNs are bipolar neurons with dendrites facing the external surface of the cribriform plate with axons that pass through the cribriform foramina with terminal end at olfactory bulbs. The ORNs are located in the olfactory epithelium in the nasal cavity. The cell bodies of the ORNs are distributed among all three of the stratified layers of the olfactory epithelium.[4]
Many tiny hair-like non-motile cilia protrude from the olfactory receptor cell's dendrites. The dendrites extend to the olfactory epithelial surface and each ends in a dendritic knob from which around 20 to 35 cilia protrude. The cilia have a length of up to 100 micrometres and with the cilia from other dendrites form a meshwork in the olfactory mucus.[5] The surface of the cilia is covered with olfactory receptors, a type of G protein-coupled receptor. Each olfactory receptor cell expresses only one type of olfactory receptor (OR), but many separate olfactory receptor cells express ORs which bind the same set of odors. The axons of olfactory receptor cells which express the same OR converge to form glomeruli in the olfactory bulb.[6]
Function
[edit]ORs, which are located on the membranes of the cilia have been classified as a complex type of ligand-gated metabotropic channels.[7] There are approximately 1000 different genes that code for the ORs, making them the largest gene family. An odorant will dissolve into the mucus of the olfactory epithelium and then bind to an OR. ORs can bind to a variety of odor molecules, with varying affinities. The difference in affinities causes differences in activation patterns resulting in unique odorant profiles.[8][9] The activated OR in turn activates the intracellular G-protein, GOLF (GNAL), adenylate cyclase and production of cyclic AMP (cAMP) opens ion channels in the cell membrane, resulting in an influx of sodium and calcium ions into the cell, and an efflux of chloride ions. This influx of positive ions and efflux of negative ions causes the neuron to depolarize, generating an action potential.

Desensitization
[edit]The olfactory receptor neuron has a fast working negative feedback response upon depolarization. When the neuron is depolarizing, the CNG ion channel is open allowing sodium and calcium to rush into the cell. The influx of calcium begins a cascade of events within the cell. Calcium first binds to calmodulin to form CaM. CaM will then bind to the CNG channel and close it, stopping the sodium and calcium influx.[10] CaMKII will be activated by the presence of CaM, which will phosphorylate ACIII and reduce cAMP production.[11] CaMKII will also activate phosphodiesterase, which will then hydrolyze cAMP.[12] The effect of this negative feedback response inhibits the neuron from further activation when another odor molecule is introduced.
Number of distinguishable odors
[edit]A widely publicized study suggested that humans can detect more than one trillion different odors.[13] This finding has been disputed. Critics argued that the methodology used for the estimation was fundamentally flawed, showing that applying the same argument for better-understood sensory modalities, such as vision or audition, leads to wrong conclusions.[14] Other researchers have also showed that the result is extremely sensitive to the precise details of the calculation, with small variations changing the result over dozens of orders of magnitude, possibly going as low as a few thousand.[15] The authors of the original study have argued that their estimate holds as long as it is assumed that odor space is sufficiently high-dimensional.[16]
Other animals
[edit]This section needs expansion. You can help by adding to it. (February 2018) |
See also
[edit]References
[edit]- ^ Berkowicz, D. A.; Trombley, P. Q.; Shepherd, G. M. (1994). "Evidence for glutamate as the olfactory receptor cell neurotransmitter". Journal of Neurophysiology. 71 (6): 2557–61. doi:10.1152/jn.1994.71.6.2557. PMID 7931535.
- ^ Vermeulen, A; Rospars, J. P. (1998). "Dendritic integration in olfactory sensory neurons: A steady-state analysis of how the neuron structure and neuron environment influence the coding of odor intensity". Journal of Computational Neuroscience. 5 (3): 243–66. doi:10.1023/A:1008826827728. PMID 9663551. S2CID 19598225.
- ^ Saladin, Kenneth (2012). Anatomy & physiology : the unity of form and function (6th ed.). McGraw-Hill. p. 593. ISBN 978-0073378251.
- ^ Cunningham, A.M.; Manis, P.B.; Reed, R.R.; Ronnett, G.V. (1999). "Olfactory receptor neurons exist as distinct subclasses of immature and mature cells in primary culture". Neuroscience. 93 (4): 1301–12. doi:10.1016/s0306-4522(99)00193-1. PMID 10501454. S2CID 23634746.
- ^ McClintock, TS; Khan, N; Xie, C; Martens, JR (5 December 2020). "Maturation of the Olfactory Sensory Neuron and Its Cilia". Chemical Senses. 45 (9): 805–822. doi:10.1093/chemse/bjaa070. PMC 8133333. PMID 33075817.
- ^ McEwen, D. P (2008). "Olfactory cilia: our direct neuronal connection to the external world". Curr. Top. Dev. Biol. Current Topics in Developmental Biology. 85: 333–370. doi:10.1016/S0070-2153(08)00812-0. ISBN 9780123744531. PMID 19147011.
- ^ Touhara, Kazushige (2009). "Insect Olfactory Receptor Complex Functions as a Ligand-gated Ionotropic Channel". Annals of the New York Academy of Sciences. 1170 (1): 177–80. Bibcode:2009NYASA1170..177T. doi:10.1111/j.1749-6632.2009.03935.x. PMID 19686133. S2CID 6336906.
- ^ Bieri, S.; Monastyrskaia, K; Schilling, B (2004). "Olfactory Receptor Neuron Profiling using Sandalwood Odorants". Chemical Senses. 29 (6): 483–7. doi:10.1093/chemse/bjh050. PMID 15269120.
- ^ Fan, Jinhong; Ngai, John (2001). "Onset of Odorant Receptor Gene Expression during Olfactory Sensory Neuron Regeneration". Developmental Biology. 229 (1): 119–27. doi:10.1006/dbio.2000.9972. PMID 11133158.
- ^ Bradley, J; Reuter, D; Frings, S (2001). "Facilitation of calmodulinmediated odor adaptation by cAMP-gated channel subunits". Science. 294 (5549): 2176–2178. Bibcode:2001Sci...294.2176B. doi:10.1126/science.1063415. PMID 11739960. S2CID 13357941.
- ^ Wei, J; Zhao, AZ; Chan, GC; Baker, LP; Impey, S; Beavo, JA; Storm, DR (1998). "Phosphorylation and inhibition of olfactory adenylyl cyclase by CaM kinase II in Neurons: a mechanism for attenuation of olfactory signals". Neuron. 21 (3): 495–504. doi:10.1016/s0896-6273(00)80561-9. PMID 9768837. S2CID 9860137.
- ^ Yan, C; Zhao, AZ; Bentley, JK; Loughney, K; Ferguson, K; Beavo, JA (1995). "Molecular cloning and characterization of a calmodulin-dependent phosphodiesterase enriched in olfactory sensory neurons". Proc Natl Acad Sci USA. 92 (21): 9677–9681. Bibcode:1995PNAS...92.9677Y. doi:10.1073/pnas.92.21.9677. PMC 40865. PMID 7568196.
- ^ Bushdid, C.; Magnasco, M. O.; Vosshall, L. B.; Keller, A. (2014). "Humans Can Discriminate More than 1 Trillion Olfactory Stimuli". Science. 343 (6177): 1370–2. Bibcode:2014Sci...343.1370B. doi:10.1126/science.1249168. PMC 4483192. PMID 24653035.
- ^ Meister, Markus (2015). "On the dimensionality of odor space". eLife. 4: e07865. doi:10.7554/eLife.07865. PMC 4491593. PMID 26151672.
- ^ Gerkin, Richard C.; Castro, Jason B. (2015). "The number of olfactory stimuli that humans can discriminate is still unknown". eLife. 4: e08127. doi:10.7554/eLife.08127. PMC 4491703. PMID 26151673.
- ^ Magnasco, Marcelo O.; Keller, Andreas; Vosshall, Leslie B. (2015). "On the dimensionality of olfactory space". doi:10.1101/022103.
{{cite journal}}: Cite journal requires|journal=(help)
